#backed up enough times to contain several terabytes worth of data
Explore tagged Tumblr posts
Text

#sev.screams#this is abt zzz btw#thinking about how android!reader will eventually outlive grace#unchanging metal against the transicence of flesh and bone#android!reader continually watching over belobog industries in grace’s honor#maybe a few hundred years in the future their parts will finally corrode#their wires snapping and their joints beginning to creak#and the new generation of engineers following in the footsteps of their predecessors#find an old android in a tomb of cables and silicone surrounded by old tv screens#the android has powered off. seemingly for the last time#the engineering team recover a data chip; perhaps there is useful information from those who came before#instead#all they find is a video of a woman smiling broadly#backed up enough times to contain several terabytes worth of data#i dont know where im going with this. just yet again obsessed w the idea of something that#was not built for the intention of loving but loves anyway. as best as they can#ursgagshdjdjsh i will never beat the hopeless romantic allegations
17 notes
·
View notes
Text
Storing data in DNA brings nature into the digital universe
by Luis Ceze and Karin Strauss

Humanity is producing data at an unimaginable rate, to the point that storage technologies can’t keep up. Every five years, the amount of data we’re producing increases 10-fold, including photos and videos. Not all of it needs to be stored, but manufacturers of data storage aren’t making hard drives and flash chips fast enough to hold what we do want to keep. Since we’re not going to stop taking pictures and recording movies, we need to develop new ways to save them.
Over millennia, nature has evolved an incredible information storage medium – DNA. It evolved to store genetic information, blueprints for building proteins, but DNA can be used for many more purposes than just that. DNA is also much denser than modern storage media: The data on hundreds of thousands of DVDs could fit inside a matchbox-size package of DNA. DNA is also much more durable – lasting thousands of years – than today’s hard drives, which may last years or decades. And while hard drive formats and connection standards become obsolete, DNA never will, at least so long as there’s life.
The idea of storing digital data in DNA is several decades old, but recent work from Harvard and the European Bioinformatics Institute showed that progress in modern DNA manipulation methods could make it both possible and practical today. Many research groups, including at the ETH Zurich, the University of Illinois at Urbana-Champaign and Columbia University are working on this problem. Our own group at the University of Washington and Microsoft holds the world record for the amount of data successfully stored in and retrieved from DNA – 200 megabytes.
Preparing bits to become atoms
Traditional media like hard drives, thumb drives or DVDs store digital data by changing either the magnetic, electrical or optical properties of a material to store 0s and 1s.
To store data in DNA, the concept is the same, but the process is different. DNA molecules are long sequences of smaller molecules, called nucleotides – adenine, cytosine, thymine and guanine, usually designated as A, C, T and G. Rather than creating sequences of 0s and 1s, as in electronic media, DNA storage uses sequences of the nucleotides.
There are several ways to do this, but the general idea is to assign digital data patterns to DNA nucleotides. For instance, 00 could be equivalent to A, 01 to C, 10 to T and 11 to G. To store a picture, for example, we start with its encoding as a digital file, like a JPEG. That file is, in essence, a long string of 0s and 1s. Let’s say the first eight bits of the file are 01111000; we break them into pairs – 01 11 10 00 – which correspond to C-G-T-A. That’s the order in which we join the nucleotides to form a DNA strand.
Digital computer files can be quite large – even terabytes in size for large databases. But individual DNA strands have to be much shorter – holding only about 20 bytes each. That’s because the longer a DNA strand is, the harder it is to build chemically.
So we need to break the data into smaller chunks, and add to each an indicator of where in the sequence it falls. When it’s time to read the DNA-stored information, that indicator will ensure all the chunks of data stay in their proper order.
Now we have a plan for how to store the data. Next we have to actually do it.
Storing the data
After determining what order the letters should go in, the DNA sequences are manufactured letter by letter with chemical reactions. These reactions are driven by equipment that takes in bottles of A’s, C’s, G’s and T’s and mixes them in a liquid solution with other chemicals to control the reactions that specify the order of the physical DNA strands.
This process brings us another benefit of DNA storage: backup copies. Rather than making one strand at a time, the chemical reactions make many identical strands at once, before going on to make many copies of the next strand in the series.
Once the DNA strands are created, we need to protect them against damage from humidity and light. So we dry them out and put them in a container that keeps them cold and blocks water and light.
But stored data are useful only if we can retrieve them later.
Reading the data back
To read the data back out of storage, we use a sequencing machine exactly like those used for analysis of genomic DNA in cells. This identifies the molecules, generating a letter sequence per molecule, which we then decode into a binary sequence of 0s and 1s in order. This process can destroy the DNA as it is read – but that’s where those backup copies come into play: There are many copies of each sequence.
And if the backup copies get depleted, it is easy to make duplicate copies to refill the storage – just as nature copies DNA all the time.
At the moment, most DNA retrieval systems require reading all of the information stored in a particular container, even if we want only a small amount of it. This is like reading an entire hard drive’s worth of information just to find one email message. We have developed techniques – based on well-studied biochemistry methods – that let us identify and read only the specific pieces of information a user needs to retrieve from DNA storage.
Remaining challenges
At present, DNA storage is experimental. Before it becomes commonplace, it needs to be completely automated, and the processes of both building DNA and reading it must be improved. They are both prone to error and relatively slow. For example, today’s DNA synthesis lets us write a few hundred bytes per second; a modern hard drive can write hundreds of millions of bytes per second. An average iPhone photo would take several hours to store in DNA, though it takes less than a second to save on the phone or transfer to a computer.

These are significant challenges, but we are optimistic because all the relevant technologies are improving rapidly. Further, DNA data storage doesn’t need the perfect accuracy that biology requires, so researchers are likely to find even cheaper and faster ways to store information in nature’s oldest data storage system.
Luis Ceze is an Associate Professor of Computer Science and Engineering at the University of Washington. Karin Strauss is a Researcher in Computer Architecture, Microsoft Research and Affiliate Associate Professor of Computer Science and Engineering at the University of Washington.
This article was originally published on The Conversation.
44 notes
·
View notes
Text
A Layman’s Guide to Data Science. Part 3: Data Science Workflow

By now you have already gained enough knowledge and skills about Data Science and have built your first (or even your second and third) project. At this point, it is time to improve your workflow to facilitate further development process.
There is no specific template for solving any data science problem (otherwise you’d see it in the first textbook you come across). Each new dataset and each new problem will lead to a different roadmap. However, there are similar high-level steps in many different projects.
In this post, we offer a clean workflow that can be used as a basis for data science projects. Every stage and step in it, of course, can be addressed on its own and can even be implemented by different specialists in larger-scale projects.
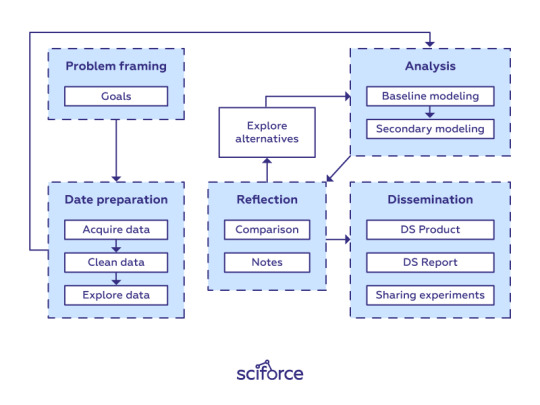
Data science workflow
Framing the problem and the goals
As you already know, at the starting point, you’re asking questions and trying to get a handle on what data you need. Therefore, think of the problem you are trying to solve. What do you want to learn more about? For now, forget about modeling, evaluation metrics, and data science-related things. Clearly stating your problem and defining goals are the first step to providing a good solution. Without it, you could lose the track in the data-science forest.
Data Preparation Phase
In any Data Science project, getting the right kind of data is critical. Before any analysis can be done, you must acquire the relevant data, reformat it into a form that is amenable to computation and clean it.

Acquire data
The first step in any data science workflow is to acquire the data to analyze. Data can come from a variety of sources:
imported from CSV files from your local machine;
queried from SQL servers;
stripped from online repositories such as public websites;
streamed on-demand from online sources via an API;
automatically generated by physical apparatus, such as scientific lab equipment attached to computers;
generated by computer software, such as logs from a webserver.
In many cases, collecting data can become messy, especially if the data isn’t something people have been collecting in an organized fashion. You’ll have to work with different sources and apply a variety of tools and methods to collect a dataset.
There are several key points to remember while collecting data:
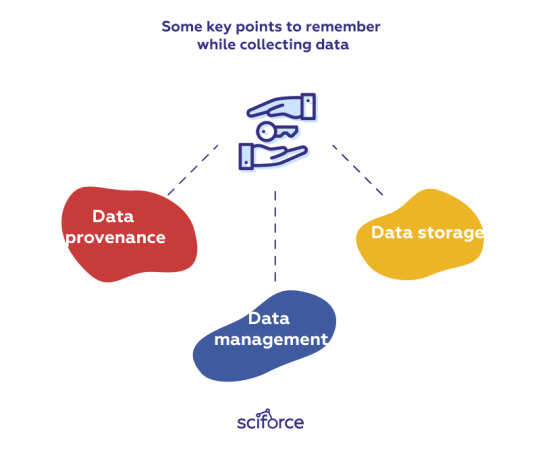
Data provenance: It is important to accurately track provenance, i.e. where each piece of data comes from and whether it is still up-to-date, since data often needs to be re-acquired later to run new experiments. Re-acquisition can be helpful if the original data sources get updated or if researchers want to test alternate hypotheses. Besides, we can use provenance to trace back downstream analysis errors to the original data sources.
Data management: To avoid data duplication and confusion between different versions, it is critical to assign proper names to data files that they create or download and then organize those files into directories. When new versions of those files are created, corresponding names should be assigned to all versions to be able to keep track of their differences. For instance, scientific lab equipment can generate hundreds or thousands of data files that scientists must name and organize before running computational analyses on them.
Data storage: With modern almost limitless access to data, it often happens that there is so much data that it cannot fit on a hard drive, so it must be stored on remote servers. While cloud services are gaining popularity, a significant amount of data analysis is still done on desktop machines with data sets that fit on modern hard drives (i.e., less than a terabyte).

Reformat and clean data
Raw data is usually not in a convenient format to run an analysis, since it was formatted by somebody else without that analysis in mind. Moreover, raw data often contains semantic errors, missing entries, or inconsistent formatting, so it needs to be “cleaned” prior to analysis.
Data wrangling (munging) is the process of cleaning data, putting everything together into one workspace, and making sure your data has no faults in it. It is possible to reformat and clean the data either manually or by writing scripts. Getting all of the values in the correct format can involve stripping characters from strings, converting integers to floats, or many other things. Afterwards, it is necessary to deal with missing values and null values that are common in sparse matrices. The process of handling them is called missing data imputation where the missing data are replaced with substituted data.
Data integration is a related challenge, since data from all sources needs to be integrated into a central MySQL relational database, which serves as the master data source for his analyses.
Usually it consumes a lot of time and cannot be fully automated, but at the same time, it can provide insights into the data structure and quality as well as the models and analyses that might be optimal to apply.

Explore the data
Here’s where you’ll start getting summary-level insights of what you’re looking at, and extracting the large trends. At this step, there are three dimensions to explore: whether the data imply supervised learning or unsupervised learning? Is this a classification problem or is it a regression problem? Is this a prediction problem or an inference problem? These three sets of questions can offer a lot of guidance when solving your data science problem.
There are many tools that help you understand your data quickly. You can start with checking out the first few rows of the data frame to get the initial impression of the data organization. Automatic tools incorporated in multiple libraries, such as Pandas’ .describe(), can quickly give you the mean, count, standard deviation and you might already see things worth diving deeper into. With this information you’ll be able to determine which variable is our target and which features we think are important.
Analysis Phase
Analysis is the core phase of data science that includes writing, executing, and refining computer programs to analyze and obtain insights from the data prepared at the previous phase. Though there are many programming languages for data science projects ranging from interpreted “scripting” languages such as Python, Perl, R, and MATLAB to compiled ones such as Java, C, C++, or even Fortran, the workflow for writing analysis software is similar across the languages.
As you can see, analysis is a repeated iteration cycle of editing scripts or programs, executing to produce output files, inspecting the output files to gain insights and discover mistakes, debugging, and re-editing.

Baseline Modeling
As a data scientist, you will build a lot of models with a variety of algorithms to perform different tasks. At the first approach to the task, it is worthwhile to avoid advanced complicated models, but to stick to simpler and more traditional linear regression for regression problems and logistic regression for classification problems as a baseline upon which you can improve.
At the model preprocessing stage you can separate out features from dependent variables, scale the data, and use a train-test-split or cross validation to prevent overfitting of the model — the problem when a model too closely tracks the training data and doesn’t perform well with new data.
With the model ready, it can be fitted on the training data and tested by having it predict y values for the X_test data. Finally, the model is evaluated with the help of metrics that are appropriate for the task, such as R-squared for regression problems and accuracy or ROC-AUC scores for classification tasks.

Secondary Modeling
Now it is time to go into a deeper analysis and, if necessary, use more advanced models, such as, for example neural networks, XGBoost, or Random Forests. It is important to remember that such models can initially render worse results than simple and easy-to-understand models due to a small dataset that cannot provide enough data or to the collinearity problem with features providing similar information.
Therefore, the key task of the secondary modeling step is parameter tuning. Each algorithm has a set of parameters you can optimize. Parameters are the variables that a machine learning technique uses to adjust to the data. Hyperparameters that are the variables that govern the training process itself, such as the number of nodes or hidden layers in a neural network, are tuned by running the whole training job, looking at the aggregate accuracy, and adjusting.
Reflection Phase
Data scientists frequently alternate between the analysis and reflection phases: whereas the analysis phase focuses on programming, the reflection phase involves thinking and communicating about the outputs of analyses. After inspecting a set of output files, a data scientist, or a group of data scientists can make comparisons between output variants and explore alternative paths by adjusting script code and/or execution parameters. Much of the data analysis process is trial-and-error: a scientist runs tests, graphs the output, reruns them, graphs the output and so on. Therefore, graphs are the central comparison tool that can be displayed side-by-side on monitors to visually compare and contrast their characteristics. A supplementary tool is taking notes, both physical and digital to keep track of the line of thought and experiments.
Communication Phase
The final phase of data science is disseminating results either in the form of a data science product or as written reports such as internal memos, slideshow presentations, business/policy white papers, or academic research publications.
A data science product implies getting your model into production. In most companies, data scientists will be working with the software engineering team to write the production code. The software can be used both to reproduce the experiments or play with the prototype systems and as an independent solution to tackle a known issue on the market, like, for example, assessing the risk of financial fraud.
Alternatively to the data product, you can create a data science report. You can showcase your results with a presentation and offer a technical overview on the process. Remember to keep your audience in mind: go into more detail if presenting to fellow data scientists or focus on the findings if you address the sales team or executives. If your company allows publishing the results, it is also a good opportunity to have feedback from other specialists. Additionally, you can write a blog post and push your code to GitHub so the data science community can learn from your success. Communicating your results is an important part of the scientific process, so this phase should not be overlooked.
0 notes
Text
Storing data in DNA brings nature into the digital universe
http://bit.ly/2vHpEiB
The next frontier of data storage: DNA. ymgerman/Shutterstock.com
Humanity is producing data at an unimaginable rate, to the point that storage technologies can’t keep up. Every five years, the amount of data we’re producing increases 10-fold, including photos and videos. Not all of it needs to be stored, but manufacturers of data storage aren’t making hard drives and flash chips fast enough to hold what we do want to keep. Since we’re not going to stop taking pictures and recording movies, we need to develop new ways to save them.
Over millennia, nature has evolved an incredible information storage medium – DNA. It evolved to store genetic information, blueprints for building proteins, but DNA can be used for many more purposes than just that. DNA is also much denser than modern storage media: The data on hundreds of thousands of DVDs could fit inside a matchbox-size package of DNA. DNA is also much more durable – lasting thousands of years – than today’s hard drives, which may last years or decades. And while hard drive formats and connection standards become obsolete, DNA never will, at least so long as there’s life.
The idea of storing digital data in DNA is several decades old, but recent work from Harvard and the European Bioinformatics Institute showed that progress in modern DNA manipulation methods could make it both possible and practical today. Many research groups, including at the ETH Zurich, the University of Illinois at Urbana-Champaign and Columbia University are working on this problem. Our own group at the University of Washington and Microsoft holds the world record for the amount of data successfully stored in and retrieved from DNA – 200 megabytes.
Preparing bits to become atoms
Traditional media like hard drives, thumb drives or DVDs store digital data by changing either the magnetic, electrical or optical properties of a material to store 0s and 1s.
To store data in DNA, the concept is the same, but the process is different. DNA molecules are long sequences of smaller molecules, called nucleotides – adenine, cytosine, thymine and guanine, usually designated as A, C, T and G. Rather than creating sequences of 0s and 1s, as in electronic media, DNA storage uses sequences of the nucleotides.
There are several ways to do this, but the general idea is to assign digital data patterns to DNA nucleotides. For instance, 00 could be equivalent to A, 01 to C, 10 to T and 11 to G. To store a picture, for example, we start with its encoding as a digital file, like a JPEG. That file is, in essence, a long string of 0s and 1s. Let’s say the first eight bits of the file are 01111000; we break them into pairs – 01 11 10 00 – which correspond to C-G-T-A. That’s the order in which we join the nucleotides to form a DNA strand.
Digital computer files can be quite large – even terabytes in size for large databases. But individual DNA strands have to be much shorter – holding only about 20 bytes each. That’s because the longer a DNA strand is, the harder it is to build chemically.
So we need to break the data into smaller chunks, and add to each an indicator of where in the sequence it falls. When it’s time to read the DNA-stored information, that indicator will ensure all the chunks of data stay in their proper order.
Now we have a plan for how to store the data. Next we have to actually do it.
Storing the data
After determining what order the letters should go in, the DNA sequences are manufactured letter by letter with chemical reactions. These reactions are driven by equipment that takes in bottles of A’s, C’s, G’s and T’s and mixes them in a liquid solution with other chemicals to control the reactions that specify the order of the physical DNA strands.
This process brings us another benefit of DNA storage: backup copies. Rather than making one strand at a time, the chemical reactions make many identical strands at once, before going on to make many copies of the next strand in the series.
Once the DNA strands are created, we need to protect them against damage from humidity and light. So we dry them out and put them in a container that keeps them cold and blocks water and light.
But stored data are useful only if we can retrieve them later.
Reading the data back
To read the data back out of storage, we use a sequencing machine exactly like those used for analysis of genomic DNA in cells. This identifies the molecules, generating a letter sequence per molecule, which we then decode into a binary sequence of 0s and 1s in order. This process can destroy the DNA as it is read – but that’s where those backup copies come into play: There are many copies of each sequence.
And if the backup copies get depleted, it is easy to make duplicate copies to refill the storage – just as nature copies DNA all the time.
At the moment, most DNA retrieval systems require reading all of the information stored in a particular container, even if we want only a small amount of it. This is like reading an entire hard drive’s worth of information just to find one email message. We have developed techniques – based on well-studied biochemistry methods – that let us identify and read only the specific pieces of information a user needs to retrieve from DNA storage.
Remaining challenges
At present, DNA storage is experimental. Before it becomes commonplace, it needs to be completely automated, and the processes of both building DNA and reading it must be improved. They are both prone to error and relatively slow. For example, today’s DNA synthesis lets us write a few hundred bytes per second; a modern hard drive can write hundreds of millions of bytes per second. An average iPhone photo would take several hours to store in DNA, though it takes less than a second to save on the phone or transfer to a computer.
These are significant challenges, but we are optimistic because all the relevant technologies are improving rapidly. Further, DNA data storage doesn’t need the perfect accuracy that biology requires, so researchers are likely to find even cheaper and faster ways to store information in nature’s oldest data storage system.
Luis Ceze works for University of Washington and consults for Microsoft Research. He receives funding from Microsoft, NSF and DARPA.
Karin Strauss works for Microsoft Research and is an affiliate faculty member at University of Washington. She is also a member of the Institute of Electrical and Electronics Engineers (IEEE), a member of the Association for Computing Machinery (ACM), and an Executive Committee member of the ACM's Special Interest Group on Computer Architecture (SIGARCH).
0 notes